Managing multi-dimensional datasets with metadata in Python
What many datasets for machine learning tutorials are lacking is metadata. Example datasets are, with good reason, kept as simple as possible and only capture the core of the problem. In the real world however, a business goal very rarely comes as one neat pack of uniform data. You may have CT images obtained with different scanners, manufacturing data from different facilities, sales data including different marketing campaigns… When working on a particular task, you want to be aware of such information, as disregarding it may lead to unexpected model behavior.
Detecting unintended bias is a crucial topic in itself. In this post however, I want to investigate how to write clean and efficient code for handling metadata alongside an actual dataset. A few tricks and design choices make working with metadata a lot easier and can facilitate a more thorough model evaluation.
The concepts described in this article do not apply to all machine learning problems. When working with tabular data, Pandas provides all necessary tools out of the box. The main challenge comes with handling multidimensional data (images, audio, etc.), which does not fit into a data frame, in combination with additional metadata, which does.
Also, this post is mainly concerning small-ish datasets, that do not run into memory limitations. For larger-scale applications, other considerations such as distributed computing and storage come into play, that overshadow the issues addressed here.
Finally, we are not really looking at problems, where both tabular and multidimensional data are used as input to a machine learning model. Here is an article on Medium highlighting that topic.
Multiple parallel arrays of data are difficult to maintain
Most tutorials and example scripts for machine learning solutions contain lines
similar to the following, which result in a set of data arrays holding the X
and y data.
|
|
Often, x_data is a multidimensional array with the input
values and y_data is a one-dimensional vector of targets.
These are the ready-to-use datasets that let you focus on the actual tutorial
content rather than cumbersome logistics.
You can simply use the arrays with model.fit(x_train, y_train).
Now, consider the following scenario: You are working on a machine learning model for a multidimensional dataset like Fashion-MNIST. In addition to the actual data, you also have some additional information associated with each sample (i.e. each image). For the Fashion-MNIST example, this could be the release date or the designated season of each piece of clothing, or even the name of the designer. Sticking to the style seen in so many tutorials, you use different arrays to store the corresponding metadata. To illustrate this, I am creating some random metadata for the Fashion-MNIST dataset:
|
|
During your experiments, you decide to only include data after a certain year for model training and start off like this:
|
|
For evaluation of the model, data is also filtered:
|
|
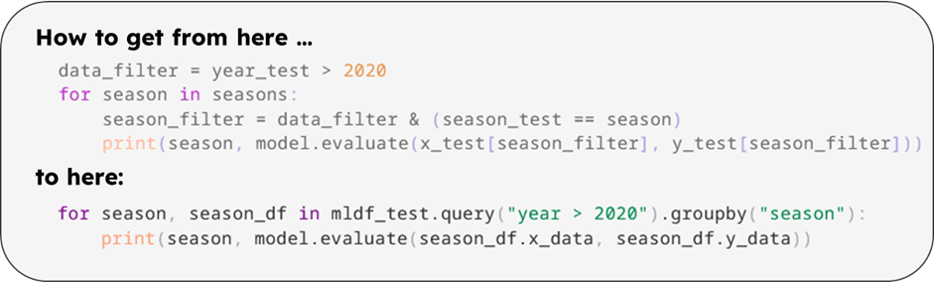
This works well with a single condition for subdividing the data. But the code gets out of hand quickly, if you want to add more filters. Let’s say you want to test the model performance separately for each design season, and write a loop:
|
|
In the last example, we at least defined each condition as a reusable boolean array. This reduces the amount of duplicated code, but it is still quite cumbersome and difficult to maintain. Wouldn’t it be nice if we could use some Pandas features for handling the data? Unfortunately, Pandas is not built to handle multidimensional arrays. So what other options do you have?
Potential solutions for handling ML datasets with metadata
The following sections suggest different approaches to moving away from multiple arrays holding data and metadata to more maintainable and readable solutions.
1) Pandas data frame with metadata and sample indices
While multidimensional ML inputs do not fit into a Pandas data frame, metadata
typically does. For each sample in x_data you have a number of attributes
(year and season in the example earlier).
We can create a data frame with the metadata. Then, we can utilize the internal
numerical index which should map 1:1 to the samples in x_data and y_data.
|
|
With Pandas, the code becomes shorter, more readable and easier to maintain.
The internal index is preserved in most Pandas operations, so the
per-season evaluation from before could now be written with a groupby.
|
|
However, with data_test, x_test and y_test, we still need to keep track
of three different objects.
With the next solution, once initialized, there is a single data frame to
deal with.
2) Subclassing Pandas data frame to include input and target data
Although it is not the most straightforward solution, we can create a custom
class based on a data frame, that can keep track of ML inputs (x_data) and
targets (y_data) while providing the ease of use given by Pandas.
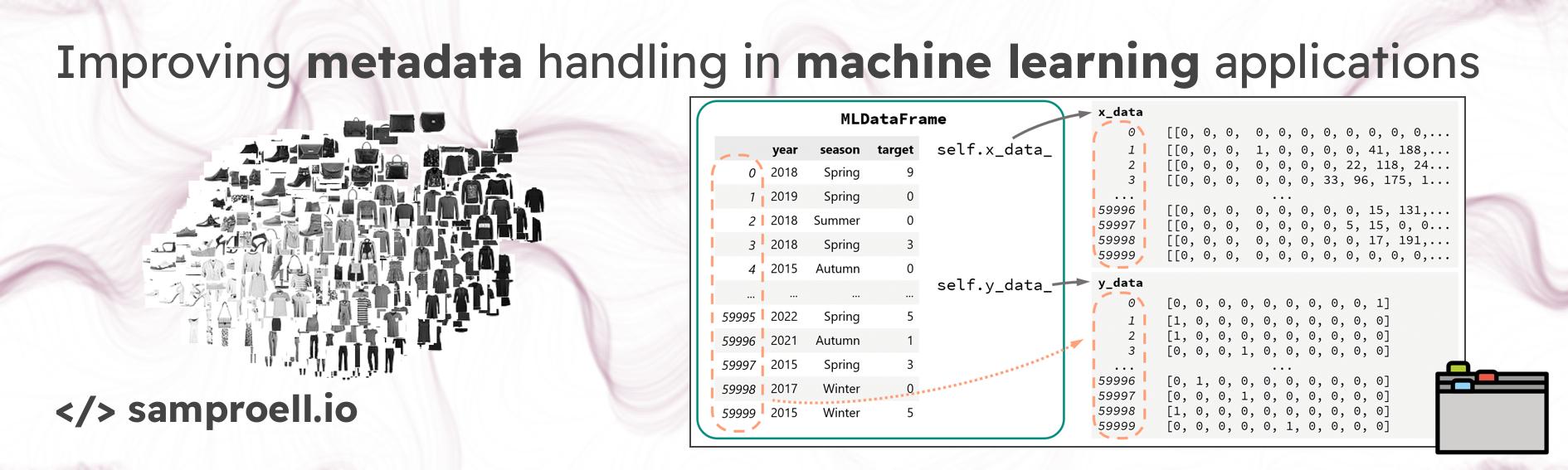
In action, such a MLDataFrame could work as follows. One can utilize many
features provided by pandas to filter/group the data and easily access the
corresponding inputs and targets for the ML task.
|
|
Let’s see how we get there.
The documentation describes the necessary steps to
subclass data structures, which is surprisingly easy.
Obviously, pd.DataFrame must be specified as the base class.
Besides, if we want inherited methods to return the modified class, a special
_constructor function must be defined.
Finally, any class attributes that should be retained by the constructor have
to be declared in the _metadata attribute. Here is everything we need,
to properly subclass a data frame.
|
|
Now, we can define our own initializer. We are giving up on a couple of positional arguments from the base initializer. Setting the index manually through the constructor is not allowed as we rely on the built-in numeric index.
|
|
We store the x and y data in private class attributes. These will always hold
the full dataset, regardless of any splitting or sub-setting of the MLDataFrame.
Two properties, aptly named x_data and y_data, then provide the actual
(potentially filtered) data arrays.
|
|
Some more adaptations and checks would be necessary, to handle bad user input
and unintended use (for example, set_index would need to be disallowed).
But as long as a user does not try to do too fancy stuff, the bare minimum
class definition above is enough.
Creating an MLDataFrame is almost like creating a regular data frame.
In this case, I am providing the metadata as a dictionary of vectors.
Additionally, the ML inputs x_train and targets y_train are given to the
data frame.
Here is a visualization of what we have achieved:

Sub-setting the data for training and evaluation becomes very clean.
|
|
A group-by-loop is now even more concise:
|
|
As a nice bonus, the MLDataFrame is compatible with scikit-learn’s most
important function: train_test_split.
|
|
Note that after filtering an MLDataFrame, the underlying x and y data stays
the same.
In fact, x_data and y_data are given as references to the the constructor
and mldf._x_data is therefore exactly the same as x_train.
However, due to numpy’s advanced indexing, the
mldf.x_data and mldf.y_data properties always return copies of the indexed
array.
|
|
3) Sticking to the data arrays and other ML dataset solutions
If you are not actually dealing with a lot of metadata or you are not required to cycle through different subsets of the same data for training and testing, it is simplest to just stick to the original format with separate data arrays. There is no need to over-engineer the metadata issue, as long as it does not get out of hand.
Alternatively, one may consider using existing dataset solutions, that
address other challenges in data handling.
In PyTorch for example, you can define
a custom DataLoader to map files on disk directly
into a dataset object for training/testing and even do some more complicated
stuff.
Tensorflow’s tf.data.Dataset can be
used to transform and augment datasets in a flexible and efficient manner.
I would also have loved to use Xarray.
But I found it to be not as intuitive and user-friendly for the metadata use
case as I was hoping.
Conclusion
Metadata plays an important role in many machine learning and data science projects. Typically, there is some nuance and context to consider when working on a particular dataset. Handling metadata in code can be tricky and if not done properly, it can result in very convoluted code that becomes error-prone and difficult to maintain. In this post, I showed a couple of options to move away from handling metadata alongside ML datasets in a more flexible and cleaner fashion.
A custom MLDataFrame class can make filtering and sub-setting data a lot less
verbose.
But using a non-standard solution for this fairly specialized problem may not
always be the best choice. The implementation described above only covers the
most simple use and would need several adaptions to improve robustness.
Also, as mentioned in the beginning, there are ML applications, where other
concerns outweigh the issues addressed with the MLDataFrame.
Nonetheless, I have found myself juggling multiple data arrays in parallel many
times, where a tool like the MLDataFrame would have helped big time.
You, also, may re-consider some of your data handling procedures, especially
when there are different scenarios and groupings to deal with.
Below is the complete MLDataFrame implementation you can copy with one click. You can find the complete example code from this post on GitHub.
|
|